One solution, exponential insight
Finally see it all
Uncover the biomarkers you’re searching for with a single workflow solution. biomodal’s multiomic analysis provides both genetic and epigenetic data simultaneously with groundbreaking clarity, precision and speed.
Genetics alone does not tell the whole story
Understanding both genetics and epigenetics with a multiomic approach can provide valuable insight into the dynamic state of a cell. Epigenetic modification of cytosine is a crucial pathway in the regulation of gene expression.
duet multiomics solution evoC
A true 6-base genome enabling the measurement of both 5‑methylcytosine (5mC) and 5‑hydroxymethylcytosine (5hmC).
duet multiomics solution +modC
This approach provides the most efficient way to obtain genetic and methylation information simultaneously.
Move beyond a 4-base genome
Our single workflow solution provides the most efficient way to obtain genetic and methylation information simultaneously from a single sample using your existing next-generation sequencing pipeline

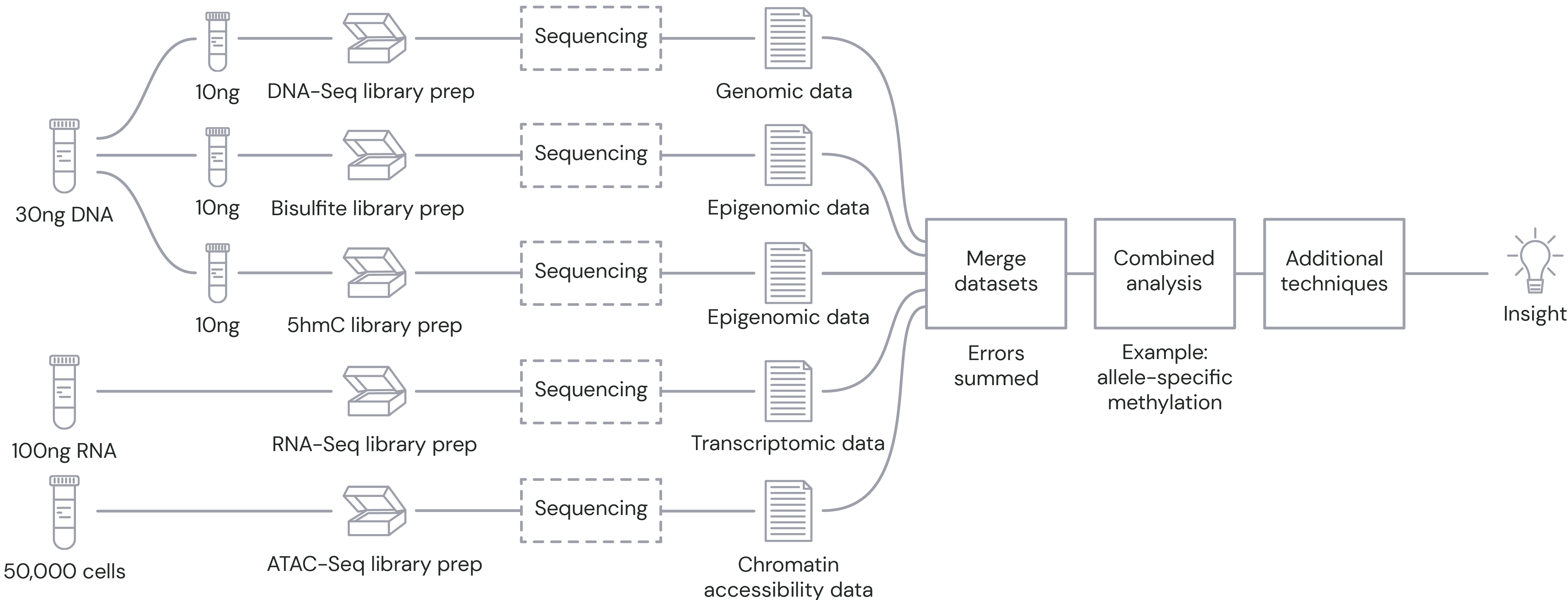
Conventional methods: multiple samples, multiple workflows, multiple data analyses
These approaches require splitting the sample and running parallel workflows, with multiple data analyses and significant information loss.
A single comprehensive workflow to uncover the DNA modifications that matter

Reveal more genetic data, all at once
High accuracy reads
The duet multiomic solution inherently suppresses PCR and sequencing errors with a base call accuracy in excess of Q40.
Seamless integration
Supercharge your genetic insights without costly in-house bioinformatics or changing your current sequencing platform.
Optimised bioinformatics
Precise sequence information in one run, eliminating the inaccuracy of inferred methylation as well as data loss from merged sequencing workflows.
Discover the full story of genetic and epigenetic change
Variant-associated methylation (VAM)
Acquiring genetic and methylation data simultaneously produces higher quality calls that show how methylation promotes or silences specific genes causing deregulation, altered expression, or potential disease.
Liquid biopsy
Maximise the genetic data you obtain from cell-free DNA (cfDNA) with our combined workflow, allowing you to use precious low input DNA samples to detect and monitor disease states.
Applications for multiomics discovery
Cancer research
Multiomics analysis offers a deeper understanding of the drivers of cancer. Our technology allows you to bring multiomic analysis to your research using your existing DNA sequencing infrastructure.
Neurodegenerative disease
Reveal the role of DNA methylation in the most common neurodegenerative diseases, and identify potential targets for promising therapeutics.
Liquid biopsy
Enhance the power of your liquid biopsy sample for disease detection and monitoring, with a low-input DNA sample.
Ageing
Probe and determine the markers of biological age to better understand age-related diseases including Alzheimer’s disease, osteoporosis, type II diabetes and chronic heart disease.
Non-invasive prenatal testing (NIPT)
Combining genetic and epigenetic DNA sequencing has the potential to reduce the time taken to detect pregnancy complications and significant genetic and epigenetic variations in a foetus.
Precision medicine
Use multiomics analysis to identify targets for promising therapeutics tailored to the genetics and epigenetics of the individual.
Explore our science
We can help you reveal new data and multimodal insights from your research.

